The following is an online supplement to the poster I’m presenting at the 2017 SEED conference in Vancouver and a talk that I gave at the 2017 EBRC conference.
What is the utility of active degradation in bacterial genetic networks and how can synthetic biology help us tease out its role in bet-hedging systems? Using a combination of synthetic reporter plasmids and mathematical modeling we investigate this problem.
As shown in this fantastic paper, even bacteria that are genetically identical can demonstrate differing susceptibility to antibiotics.
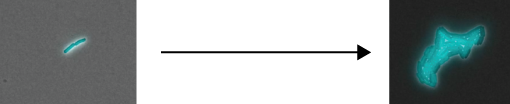
The video below shows a growing bacterial microcolony that’s hit with a temporary dose of carbenicillin. Red coloration is a propidium iodide indication of cell death. Cyan represents diversity in a transcriptional reporter for the multiple antibiotic resistance activator MarA
We see that some cells die and others live - the cells that live go on to reproduce the same diversity. Moreover we see that this survivorship is correlated with MarA level. From literature, we know that MarA is actively degraded by Lon protease. How does turnover in MarA affect the distribution of the resistance phenotype?
Consider the following distributions
Produced by the following stationary ergodic processes.
Even though the distributions have the same statistics, we see that the processes that produce each distribution are changing differently in time.
This difference can be described by the metric correlation time, which is to say how well knowing the state of the processes now informs where it will be in the future. For instance, pure white noise has a correlation time of 0 minutes - knowing where it is now does not inform at all where it will be in the future.
Additionally, thanks to the assumption of ergodicity we can assume that following one time trace for a sufficiently long period of time is the same as ob many processes at the same time point.
 Cell growth is clearly important for the development of a diverse population, but more than that the noise profile of the genes of interest is important as well.
Cell growth is clearly important for the development of a diverse population, but more than that the noise profile of the genes of interest is important as well.
We can model the development of diversity in noisy genes by an Ornstein–Uhlenbeck processes
Where \(\theta_{x}\) scales with the half-life of the molecule.
Below we see simulation of several trajectories as well as the analytical solutions to the variance over time of the function for a short half-life activator. Click links for interactive versions.
Increasing the half-life of the activator demonstrates a slower accumulation of diversity over time.
Mirroring the computational results, experimental results for activators with differing half-lives demonstrate different rates of diversity generation. Here we compare a system with wild type MarA that is actively degraded to a system with synthetically modified MarA that is not.
So far we’ve only discussed diversity in an activator, but MarA doesn’t operate in isolation, it goes on to activate multiple downstream genes to coordinate a cohesive resistance phenoptype.
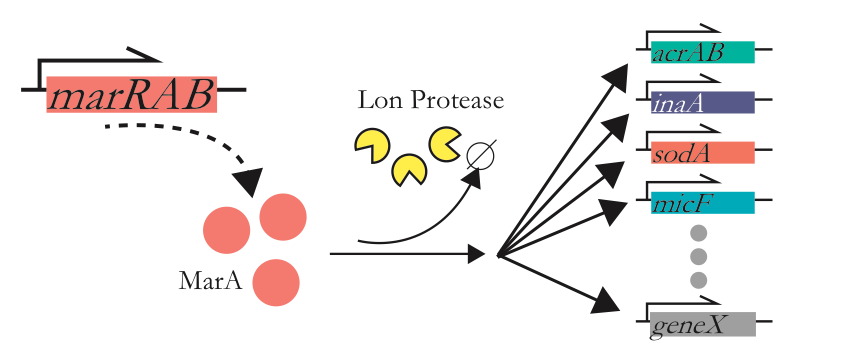
For more information on half-life may affect downstream coordination, please see the associated poster.